Trying to determine the causative agent for outbreaks of foodborne illnesses can be a gargantuan epidemiological task. It requires discerning which foods were consumed by ill people, who ate what, when and where, and finally doing the lab work to determine what agent (was it bacterial, viral or parasitic) was responsible. The time-honored way to determine if and which microbe was responsible is to grow the suspect microbe on an agar plate in the lab. This can, depending on exactly which bug one is trying to grow, take days. But newer techniques have been devised that can provide faster results, thus allowing scientists to both determine the source more quickly, or decide which antibiotics might be most effective.
These so-called culture-independent diagnostic tests, or CIDTs don’t require the suspected pathogen to be grown in the lab, and thus are faster than traditional modes of identification — a result may be obtained in hours rather than days. They can involve detecting bacterial antigens, nucleic acid sequences, toxins or toxin genes. However, although speed is really important for example in deciding which antibiotic to use, there can be some drawbacks to the newer testing techniques.
For example, definitive identification of a pathogen (say Salmonella) requires obtaining an isolate or pure sample. Then the identity is further determined to learn which of several possible subtypes, or serotypes, the suspect is. For Salmonella, it could be S. typhimurium, or S. heidelberg, or one of several others. This information can be crucial in determining which food caused the outbreak of food poisoning.
Another important question is if, or the extent to which, the use of these newer, more sensitive techniques might have an impact on the data used to track the incidence and prevalence of specific pathogens. Ellyn P. Marder from the CDC and colleagues used preliminary data from the 2017 10 U.S. sites of the Foodborne Diseases Acctive Surveillance Network to examine this possibility. Compared with the data from 2014-2016, in 2017 saw increased incidences of infections with five pathogens, as shown in the figure. 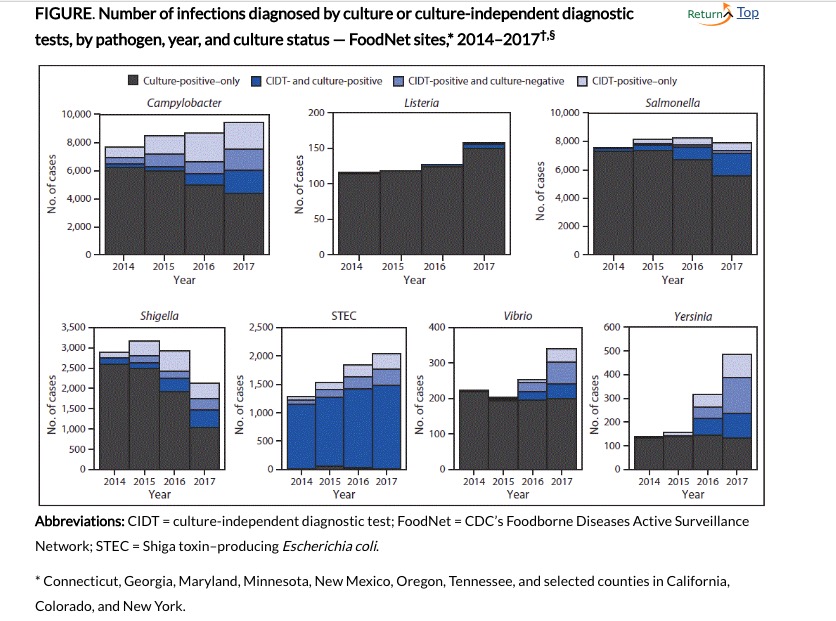 The black Source: Marder, MPH EP, Griffin PM, Cieslak PR, et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2017. MMWR Morb Mortal Wkly Rep 2018;67:324–328.
The black Source: Marder, MPH EP, Griffin PM, Cieslak PR, et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2017. MMWR Morb Mortal Wkly Rep 2018;67:324–328.
portions of the bars indicate cases determined by cultures, the dark blue were positive by both culture and CIDT testing, the lighter blue were positive by CIDT but negative by culture, and the lightest potions were CIDT positive only (not tested by culture).
Clearly, use of the CIDT tests varied with respect to the pathogens presented, as did the effects on the frequency of appearance.
When they compared the incidence of the tested pathogens with those from 2014-2016, the investigators found significantly increased incidences for Cycospora, (489 percent increase), Yersinia (166 percent), Vibrio (54 percent), STEC (Shiga-toxin E. coli by 28 percent) Listeria (26 percent), and Campylobacter (10 percent). They also found that “Bacterial infections diagnosed by CIDT increased 96% overall (range = 34%–700% per pathogen) in 2017 compared with those diagnosed during 2014–2016.” When reflex cultures were performed (i.e. culturing the CIDT positive samples), 38 percent of Vibrio to 90 percent of Salmonella were also positive.
In their discussion, the authors emphasized that
CIDTs pose challenges to public health when reflex culture is not performed. Without isolates, public health laboratories are unable to subtype pathogens, determine antimicrobial susceptibility, and detect outbreaks.
In no way were they suggesting abandoning the newer technologies, rather, they note that the Association of Public Health Laboratories recommends that a positive result on CIDT tests should be followed by culture tests to determine the actual subtype of pathogen present.
Thus, although the newer CIDT tests are faster than the older culture tests, their downside is the lack of complete information they provide. However, using these tests first could decrease the time to determine appropriate treatment, while decreasing the necessity for culturing all samples, thus saving staff time and funds spent while still providing the necessary information for accurate epidemiological determinations.




