It is rare to find a mention of Giliead's remdesivir, arguably the most promising drug in the fight against coronavirus, without also hearing that it failed miserably in fighting Ebola. Your average non-scientist (and some scientists as well) will probably think that this is an indication that it's a "weak" drug that got pulled out of the garbage in desperation and tested against coronavirus simply because there was no alternative.
This is just plain wrong.
A new paper in the Journal of Biological Chemistry explains that not only is remdesivir's failure against Ebola not a black mark against the drug but is actually a checkmark that shows that it works even better than expected, at least at the molecular level. Remdesivir inhibits the polymerase enzyme of a number of RNA viruses. Polymerase enzymes are responsible for synthesizing the RNA that is essential to form new viruses, so if a drug can inhibit the polymerase of a particular virus it should shut down the replication process (Figure 1). And remdesivir is considerably better at inhibiting the SARS-CoV-2 (coronavirus) polymerase than the Ebola polymerase, which probably explains why the drug failed as a treatment for Ebola but looks promising for treating coronavirus.
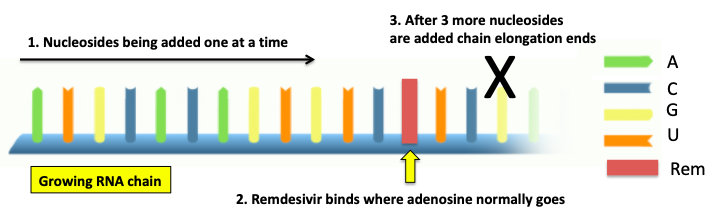
Figure 1. A simple diagram showing the inhibition of SARS-CoV-2 polymerase by remdesivir. Image: Your genome
The polymerase forms RNA by adding the nucleosides adenosine (A), cytidine (C), uridine (U), and guanosine (G) from left to right, each building block extending the newly forming RNA by one unit. But remdesivir (red rectangle), which is structurally similar to adenosine, (Figure 2) competes with for its binding site on the polymerase. If a molecule of remdesivir (1) binds to the polymerase in place of adenosine the addition process ends three nucleotides later (black X) and RNA synthesis ceases.

Figure 2. Adenosine (red oval) and remdesivir (green oval), shown in their triphosphate forms, are structurally similar, allowing remdesivir to fit into the adenosine binding site on the polymerase.
Ebola vs. SARS-CoV-2 ("coronavirus")
Both Ebola and SARS-CoV-2 have polymerase enzymes that synthesize each of the virus' genome, but the interaction between remdesivir and each of the two polymerases is substantially different (Table 1). When studying the kinetics (reaction rates) of enzymatic reactions there are certain parameters that describe how fast and tightly molecules bind to enzymes. One of the parameters used (don't make me explain this or I'll hang myself) is called Vmax/Km (orange box) and is a measure of binding efficiency, the higher the number, the better the binding (3). In the left column is ATP, the normal substrate for the polymerase. SARS-CoV-2 polymerase (red arrow) binds to adenosine triphosphate (2) (ATP) with an efficiency of 23 (red oval) while remdesivir triphosphate (RDV-TP) does so with an efficiency of 84 (red square). This means that the drug binds "better" to the polymerase by 84/23, (3.6-times) than ATP, the normal substrate. This is what you want a drug to do.
But with Ebola polymerase (blue arrow) it's a different story. In this case ATP, the natural substrate binds with an efficiency of 1.1 while this figure for the drug (remdesivir) is only 0.28. This means that the drug binds "worse" by 1.1/0.28 (4-times) than ATP. This is what you don't want a drug to do.
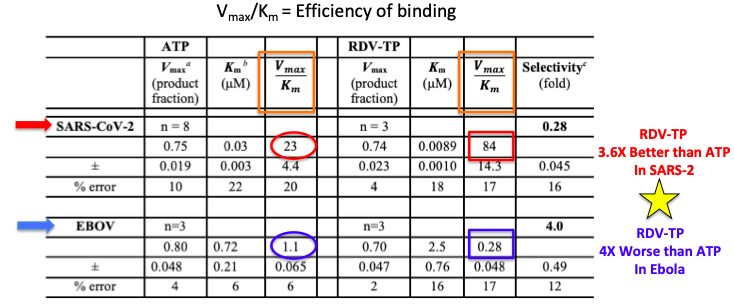
Table 1: Binding parameters of ATP and remdesivir triphosphate to the polymerases of both Ebola and SARS-CoV-2. Remdesivir binds 3.6X more effectively than ATP in SARS-CoV-2 polymerase, but 4X less effectively in Ebola polymerase. Source: JBC
What do these numbers mean?
In vitro, remdesivir is a more potent inhibitor of the SARS-CoV-2 polymerase than it is of the Ebola polymerase. In other words, remdesivir efficiently shuts SARS-CoV-2 polymerase but not Ebola polymerase. All things equal, remdesivir should inhibit the replication of down coronavirus far better than it did for Ebola.
This provides a rational explanation of why the same drug could fail to treat Ebola but succeed for coronavirus, but it is not evidence. In drug discovery things are never equal, so surprises (good and bad) are not unusual. The evidence of remdesivir's efficacy (or lack thereof) will only be seen once clinical trials are complete.
UPDATE: April 16th. It seems to be working. From STAT: Early peek at data on Gilead coronavirus drug suggests patients are responding to treatment
NOTES:
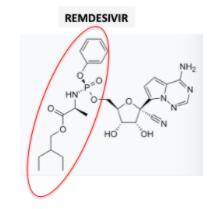
(1) Remdesivir is a pro-drug of remdesivir triphosphate. The pro-drug fragment is shown in the red oval. The cleavage of the pro-drug to the drug is an enzymatic reaction.
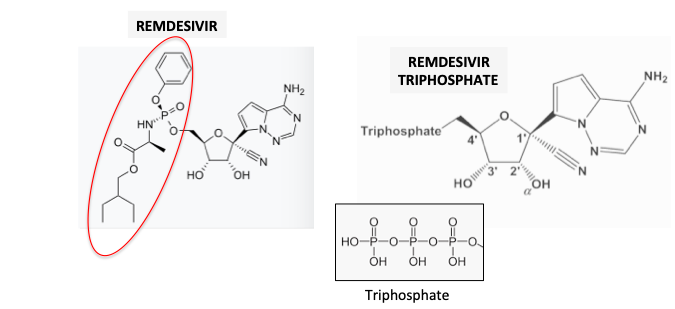
(2) ATP is widely known as the source of cellular energy. But it's also a nucleotide building block, appearing in RNA and DNA.
(3) For the deranged/bored/masochists out there:
Vmax is the speed at which the enzyme binds to a substrate. Km is the strength of that bond. The lower the number the better. These "definitions" are intentionally oversimplified.



