 Image courtesy of news.mit.edu
Image courtesy of news.mit.edu
With all of the technology on the MIT campus, it is not every day that paper is at the center of a new discovery.
This time, a team in the Department of Biological Engineering and Institute for Medical Engineering and Science, in collaboration with others, has developed a new device to detect the presence of Zika virus from biological samples.
With Zika infection, simply knowing that you have a Zika infection or are "Zika positive" could slow the rate of transmission considerably, which makes testing a lynchpin in the fight against Zika -- putting it in the same category as a treatment or a vaccine.
This is because a person who is infected can transmit the virus in two different ways that we know of. First, if an uninfected mosquito bites an infected person, the mosquito can become infected, and then pass the virus to an uninfected person. Also, there are known cases of sexual transmission from a man to his sexual partner.
So, the more infected people = the more mosquitoes carrying Zika. And, the more mosquitoes carrying Zika = more infected people.
The other reason that widespread testing is important is because many people who have Zika do not know it, because they are asymptomatic and less likely to practice safe sex or prevent mosquito bites.
There are two different methods currently being used to test for the presence of Zika now, but they are long, laborious and may not be able to distinguish between Zika and the closely related Dengue virus.
This new technology has advantages over current detection methods because it is more sensitive (can detect less virus), faster and is specific to Zika (will not mistake Zika for Dengue or other viruses). It's also less expensive and requires less work.
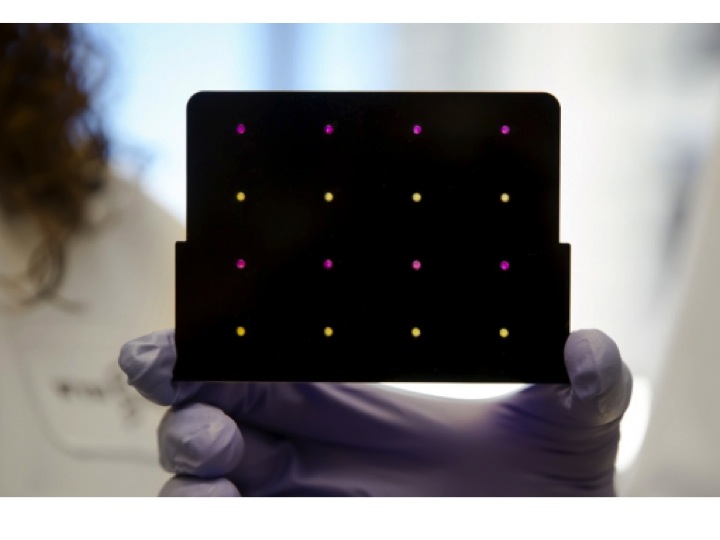
Here's how it works: Paper discs are embedded with 24 pieces of the genome of Zika virus called sensors. The sequence of the Zika genome is unique; no other virus in existence has the same sequence. It's like fingerprints are to humans.
When a blood, urine or saliva sample is added to the disc, it changes color from yellow to purple (as seen in the above photo), but only if the sample contains pieces of RNA that match the Zika RNA already on the disc. Purple means positive and yellow means negative.
There are many, many advantages to this device:
- Test results are available incredibly quickly (within a few hours)
- It can distinguish between Zika and other very similar viruses, such as Dengue.
- It can be stored at room temperature
- The results can be read by eye or with an inexpensive, battery-operated reader
- It's inexpensive
- Because of a built-in amplification system, the test can detect very low levels of virus (2 to 3 ppq, or parts per quadrillion). This is the same as one human hair out of all the hair on all the heads of all the people in the world.*
- It can work with blood, urine, or saliva samples
- Through an added step, the device can distinguish between strains of Zika virus
In other good news, the CDC reported earlier this week that Zika virus RNA is able to be detected for up to two weeks in urine, as compared to one week in blood. This is incredibly important in the fight against Zika. The extra week will allow for many more cases to be diagnosed. Also, it goes without saying that urine is easier to collect than blood.
More work and testing need to be done in order to get this device out of the lab and into the field. But the lead author on the paper, Dr. James Collins, optimistically reports that they are "not far off." And this technology can be harnessed in the future, for the next Zika-type virus that comes our way.
*http://www.waterontheweb.org/resources/conversiontables.html



